The 11 projects of the priority program SPP InterZell focuses on the cell-cell and the cell-bioreactor interactions. The "Novel production processes through cross-scale analysis, modeling and design of cell-cell and cell-bioreactor interactions (InterZell)“ offers new research approaches for biotechnical production processes. Despite intensive research, the questions of cell-cell interaction in mixed cultures or cell-bioreactor interaction remain unanswered and often lead to production losses. Questions like these will be addressed by InterZell research projects.
Project:
In this project, the researchers will develop a bioprocess design and understanding to stably and productively operate and scale filamentous co-culture fermentations for the production of natural products. Exemplarily, in the project a co-culture bioprocess for the cooperation of the cellulose degrader Trichoderma reesei, a filamentous fungus, and Streptomyces coelicolor will be developed. S. coelicolor is a filamentous bacterium producing several natural products such as actinorhodin, a quinone polyketide antibiotic, which is also used as a pH indicator or methylenomycin, a cyclopentanoid antibiotic. The quantitative spectrum of the natural products is dependent on various environmental and physiological factors. The robustness of the process design and operation will be validated by exchanging the natural product producer for other Streptomyces species. To establish a stable bioprocess strategy for the productive co-cultivation of the filamentous organisms T. reesei and S. coelicolor the engineering and microbiological approaches will be combined. Therefor a strategic bioprocess development, co –culture dynamic investigation and the establishing of a stable co-culture bioprocess for optimal cultivation of the filamentous system and the interaction between the co-cultured species will be estimated and combined.
Prof. Dr. rer. nat. Miriam Agler-Rosenbaum
Leibniz Institute for Natural Products Research and Infection Biology
Hans-Knöll-Institute (HKI) Synthetic Biotechnology
Beutenbergstrasse 11a, 07745 Jena
Prof. Dr.-Ing. Jochen Büchs
RWTH Aachen University
Aachen Verfahrenstechnik
Biochemical Engineering (AVT-BioVT)
Forckenbeckstr. 51, 52074 Aachen
Project:
Current bioprocesses for production of value-added compounds are mainly based on pure cultures that are composed of rationally engineered strains of platform organisms with versatile metabolic capacities. These strains often possess vast overcapacities of specific central metabolic genes, a natural mechanism to cope with rapidly changing environmental conditions. In the comparably well-defined environment of a bioreactor, this metabolic flexibility is much less required and results in suboptimal production processes due to the waste of energy. In this project, the researchers want to generate different genome reduced strains of the same species by targeting relevant, energy demanding pathways such as amino acid biosynthesis. Due to feedback control, enzymes in these pathways are often not active, that means a lot of energy is wasted by production of these proteins. By harnessing this overcapacity to cross-feed another strain, it is aimed to optimize resource use and thereby generate superior Communities of Niche optimized Strains (CoNoS). Starting with the first work package the strain design, construction and analysis of auxotrophic C. glutamicum strains by knock-down of whole amino acid biosynthesis pathways using CRISPRi, the strain evaluation and evolution will be tested and strain and processing optimization for final CoNoS in lab-scale operation through quantitative omics will be optimized.
Dr. Meike Baumgart
Forschungszentrum Jülich GmbH
Institut für Bio- und Geowissenschaften
IBG-1: Biotechnologie, 52425 Jülich
Dr.-Ing. Stephan Noack
Forschungszentrum Jülich GmbH
Institute of Bio- and Geosciences
Microbial Bioprocess Lab, 52425 Jülich
Project:
The objective of this project is to gain quantitative and fundamental knowledge of defined mixed cultures and to develop corresponding novel process strategies for the production of caproic acid (CA) from different carbon sources. CA is a highly appealing C6-compound with a wide range of high-value applications in agriculture, chemical and pharmaceutical industries. Presently CA is mainly produced from fossil-based n-hexanol. No more than 7 different bacterial species have been reported to be capable of CA formation through chain-elongation via reverse β oxidation. In the anticipated CA producing mixed culture one partner will produce lactate from a primary carbon source and the other partner will convert the intermediate lactate into CA. The work package contains strain construction for mixed cultures. Respective genetic potential is present in C. carboxidivorans, C. kluyveri, strain CPB6 and a new caproate producing strain termed EA1. Metabolic engineering strategies will be applied to improve the performance of the mixed cultures, especially with respect to the removal of major byproduct. Quantitative characterization of synthetic mixed cultures will be carried out, especially regarding the growth physiology and product formation kinetics, and establishment of novel fermentation process and apparatus for the cultivation and study of mixed cultures, modelling and process control strategies and assessment of the synthetic cultures in view of scale-up.
Dr. Frank Bengelsdorf
Universität Ulm
Institut für Mikrobiologie und Biotechnologie
Albert-Einstein-Allee 11, 89081 Ulm
Prof. Dr. An-Ping Zeng
Technische Universität Hamburg
Bioprozess- und Biosystemtechnik
Denickestraße 15 (K), 21073 Hamburg
Project:
Filamentous fungi including Aspergillus niger are cell factories for the production of organic acids, proteins and bioactive compounds. Traditionally, stirred-tank reactors (STRs) are used to cultivate them under highly reproducible conditions ensuring optimum oxygen uptake and high growth rates. However, the evolution of their macroscopic morphologies – ranging from freely dispersed mycelia over loose clumps to dense pellets - is an unpredictable, multifactorial process, which strongly limits productivities. Mechanical stirring causes high shear forces, thus affecting fungal physiology and macromorphologies. Besides mono-species heterogeneity of fungal cultures, co-cultivation of different filamentous organisms has also an impact on the culture’s performance and their product portfolio. To comprehensively understand the evolution of mono-species (A. niger) and multi-species (A. niger and Streptomyces coelicolor) pellet morphologies in lab-scale and large scale bioreactors, their impact on the established product portfolio of A. niger (primary metabolite citric acid, enzyme glucoamylase, secondary metabolite enniatin B) and the induction of new products (secondary metabolites) due to the presence of S. coelicolor under different biological and physical conditions. Computational fluid dynamics simulations will further allow determination of effective shear rates for reactor types. The model-based optimization towards a targeted morphology should be employed for future process developments. Validated models will support scale-up and scale–down for the processes.
Professor Dr.-Ing. Heiko Briesen
Technische Universität München
Wissenschaftszentrum Weihenstephan, Lehrstuhl für Systemverfahrenstechnik
Gregor-Mendel-Straße 4, 85354 Freising
Professorin Dr.-Ing. Vera Meyer
Technische Universität Berlin
Institut für Biotechnologie Fachgebiet
Angewandte und Molekulare Mikrobiologie
Gustav-Meyer-Allee 25, 13355 Berlin
Professor Dr. Peter Neubauer
Technische Universität Berlin
Institut für Biotechnologie Fachgebiet Bioverfahrenstechnik
Ackerstraße 76, 13355 Berlin
Project:
Synthetic co-cultures can be used to study cell-cell interactions between populations. These interactions are strongly determined by the cultivation technology used and hence needs to be carefully considered. Depending on the cultivation setup, co-cultures develop differently as the mode of mass exchange between the cell populations differs. As biological model systems for productive co-cultures, this research project will rely on existing synthetic co-cultures with relevance for industrial biotechnology. As two highly complementary co-culture systems, the researchers will employ a culture of Corynebacterium glutamicum that produces L-lysine from sucrose produced by E.coli. The obtained results with these model systems will be used as a basis to unravel the mass fluxes in co-cultures of phototrophic microbes (Synechococcus elongatus PCC7942 and Synechocystis sp. PCC6803) that supply their photosynthesis products as substrate to Corynebacterium glutamicum for producing L-lysine from CO2 and light. On the basis of these experiments, a dynamic modelling framework for describing and predicting the interactions of productive cells in co-cultures will be established. The researches will reach this goal by three interdisciplinary working lines: connecting advanced microfabrication, quantitative physiology of single cells and bioprocess engineering. A microfluidic co-cultivation platform and analysis workflow for mixed culture single-cell cultivations will be developed and applied to unravel cell-cell interactions. Its consequences for growth kinetics at single-cell level will be measured and modelled.
Dr.-Ing. Christian Dusny
Helmholtz-Zentrum für Umweltforschung - UFZ
Umwelt- und Biotechnologie Department Solare Materialien
Permoserstraße 15, 04318 Leipzig
Professor Dr.-Ing. Alexander Grünberger
Universität Bielefeld
Technische Fakultät Forschungsgruppe Multiscale Bioengineering
Postfach 100131, 33501 Bielefeld
Professor Dr. Dietrich Kohlheyer
Forschungszentrum Jülich GmbH
Institut für Bio- und Geowissenschaften (IBG) Systembiotechnologie (IBG-1)
Leo-Brandt-Straße, 52425 Jülich
Project:
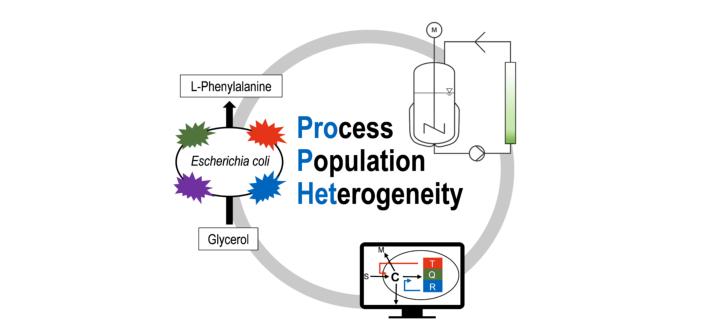
Upscaling of bioprocesses is often accompanied by loss in productivity compared to well mixed lab-scale processes. One reason is the formation of population heterogeneity. Even though this phenomenon is well studied, its mechanistic understanding is low. ProPHet is aiming for quantitative understanding of population heterogeneity originating from cell bioreactor interaction in a well-characterized fed-batch process with E. coli FUS4 strains converting glycerol to L-phenylalanine (L-Phe). To monitor population heterogeneity a quadruple reporter strain expressing four different fluorescent proteins related to growth, general stress response, oxygen limitation and L-Phe formation of single cells will be developed. The responses of the four markers will be correlated to changes in the environment of the cells. After characterization of the quadruple reporter strain in well-mixed fed-batch L-Phe production processes, the process will be transferred to a scale-down reactor to investigate the influence of gradients of process state variables on the level of population heterogeneity. The scale-down reactor will consist of a stirred-tank reactor coupled to a plug flow reactor in a by-pass. Fluorescence distributions of the quadruple strain during fed-batch processes will be collected with flow cytometry and significant subpopulations will be sorted by fluorescence activated cell sorting (FACS) and analyzed by quantitative proteomics (LC-MS/MS) for functional characterization of different phenotypes. Mathematical modeling with two approaches (population balance equations and agent based modeling) will be used to integrate the experimental data from fed-batch processes with the quadruple reporter strain into a coherent framework. After model validation and model analysis, finally, optimal conditions for production of L-Phe in fed-batch processes will be predicted and experimentally verified. With ProPHet, phenomena that lead to population heterogeneity induced variations in process performance during scale-up will be better understood. Ideally, conclusions can be drawn in which phases of the fed-batch process population heterogeneity serves a beneficial function respectively should rather be reduced.
Dr. Anna-Lena Heins
Technische Universität München
Fakultät für Maschinenwesen Lehrstuhl für Bioverfahrenstechnik
Boltzmannstraße 15, 85748 Garching
Professor Dr.-Ing. Andreas Kremling
Technische Universität München
Fakultät für Maschinenwesen Lehrstuhl für Bioverfahrenstechnik
Boltzmannstraße 15, 85748 Garching
Project:
The intrinsic capacities of photosynthetic microbes for the production of interesting compounds are limited and efficiencies are low. To overcome this bottleneck the project is using a platform in which sugar production by a photosynthetic organism (S. elongatus PCC7942) is directly coupled to the growth of a heterotrophic production strain (Pseudomonas putida EM173). The cyanobacterium fixes CO2 through photosynthesis and secretes sufficient sucrose to support the growth of P. putida, which has been genetically engineered to metabolize sucrose as the only carbon source. This engineered mixed culture allows genetically engineer P. putida in order to optimize the PHA accumulation, but also in a future scenario, to use this strain as a chassis for SynBio applications and the production of other plasmid-encoded compounds. The project aims to understand the mutual effects and influences of both microbial partners on each other and plan to enhance the performance of the system by model driven metabolic engineering. The project partners will analyze to which extend both partners are exchanging information and whether this is unidirectional or whether there is also an informational backflow from the heterotroph to the cyanobacterium. Therefore, it aims to tackle this point by following the dynamics of each sub-population and to study the effects of artificially increasing or diminishing one of them on the overall stability of the mixed-species consortium. This will be accompanied by mathematical modelling on two levels: (i) with the transcriptome data, a gene regulatory network will be set up for each species, and together with a flux model, this will lay the basis for metabolic engineering and (ii) on the population level, a dynamical model will be studied to understand interdependencies between both organisms.
Professor Dr.-Ing. Andreas Kremling
Technische Universität München
Fakultät für Maschinenwesen Lehrstuhl für Bioverfahrenstechnik
Boltzmannstraße 15, 85748 Garching
Dr. Katharina Pflüger-Grau
Technische Universität München
Fakultät für Maschinenwesen, Lehrstuhl für Bioverfahrenstechnik
Boltzmannstraße 15, 85748 Garching
Project:
Bacterial fermentation of synthesis gas (CO, H2 and CO2) is an environmental friendly approach for the synthesis of organic chemicals, which is nowadays limited to short-chain alcohols or carboxylic acids. The establishment of a stable synthetic bacterial co-culture is promising to enhance product size towards medium chain alcohols via metabolic interaction. Autotrophic growing acetogenic microorganisms such as C. carboxidivorans are capable of forming organic acids and alcohols from carbon monoxide, with ethanol and acetate as the main products. C. kluyveri, which can grow solely with acetate and ethanol as carbon sources, is able to form longer-chain organic acids like butyrate, hexanoate and octanoate (chain elongation). In this project, reaction engineering studies on both pure cultures will be done in fully controlled stirred-tank bioreactors with continuous gas supply. Kinetic studies of chain elongation and alcohol formation will identify optimal parameters for a stable synthetic co-culture. Stability of the co-culture will be monitored by flow-fluorescence in situ hybridization. Furthermore, C. kluyveri will be genetically improved concerning enhanced chain elongation capabilities and reduction properties for fatty acids into their corresponding alcohols utilizing proteomics data under co-culturing conditions. Afterwards, the autotrophic co-cultivation of C. carboxidivorans and improved C. kluyveri will be studied within the best operation conditions.
Professor Dr.-Ing. Dirk Weuster-Botz
Technische Universität München
Fakultät für Maschinenwesen Lehrstuhl für Bioverfahrenstechnik
Boltzmannstraße 15, 85748 Garching
Professor Dr. Wolfgang Liebl
Technische Universität München
Wissenschaftszentrum Weihenstephan Lehrstuhl für Mikrobiologie
Emil-Ramann-Straße 4, 85354 Freising
Project:
In biopharmaceutical engineering, the efficient and reliable transfer of processes from laboratory cultivation to production-scale bioreactors is of tremendous importance to enable the formation of new drugs with less side effects and to shorten the time from mind to market. Such a scale-up requires deep understanding of the impact of environmental conditions on cellular metabolism and regulation. The knowledge will allow quality-by-design irrespective of the cultivation scale. In this project, Particle Tracking Velocimetry and Particle Image Velocimetry as well as laser induced fluorescence will be used to investigate flow structures, streamlines, residence time distributions and concentrations fields in detail on laboratory and industrial scale, to identify compartments of relevant environmental conditions. A new measurement technique will be adopted and/or developed to follow a cell on a three-dimensional streamline in the industrial-scaled reactor, transferable to other applications. Experimental lifelines will be evaluated with respect to their putative impact on cellular performance. Parameters such as the frequency, amplitudes and duration of micro-environmental changes will be qualified, ranked and finally selected for experimental studies. Conventional scale-up simulators such as STR-PFR (stirred tank reactor – plug flow reactor) setups will be configured to mimic observed gradients of dissolved oxygen, carbon and nitrogen containing substrates or pH. Cultivations will be performed using an IgG1 producing CHO cell line of industrial relevance. Based on the experimental observations metabolic models will be identified and that can be used to predict fundamental growth kinetics and cellular energetics. Such models will be linked to bioreactor models to predict spatial inhomogeneities of large-scale production reactors. Both, compartment-based models and computational fluid dynamics will be applied to simulate large-scale conditions. Modelling will be achieved in close cooperation between the two partners exploiting the individual knowhow of preliminary microbial and engineering studies.
Professor Dr.-Ing. Ralf Takors
Universität Stuttgart
Institut für Bioverfahrenstechnik (IBVT)
Allmandring 31, 70569 Stuttgart
Professor Dr.-Ing. Michael Schlüter
Technische Universität Hamburg
Institut für Mehrphasenströmungen
Eißendorfer Straße 38, 21073 Hamburg
Project:
MiMiCry wants to exploit the approach by synthetically implementing novel properties in a consortium of two E. coli strains that necessarily can only grow and produce in symbiosis. The approach will be developed focusing on the industrially promising antibiotic/antimicrobial violacein as a show case product. Violacein is a tryptophan-derived violet pigment (water-insoluble, ethanol-soluble) with antimicrobial, antiviral, antitumoral and antiparasitic properties. MiMiCry addresses the strain engineering, process set-up and modelling: Two E. coli strains complementing each other will be engineered- producer strain I will be genetically engineered to be auxotrophic for L-tryptophan, which together with pathway improvements, leads to excretion of the intermediate, anthranilate (ANT). Producer strain II is unable to produce ANT on its own due to a trpE mutation, but is able to transform exogenous ANT to L-Trp by engineered genes. In a co-culture, strain I can feed strain II which in turn provides diffusible L-Trp to strain II. Thus, a stable growth of the culture is only possible if both strains cooperate. MiMiCry follows two conceptual setups. Non-structured, segregated models will be used to model the growth, substrate uptake and product formation kinetics, comprising substrate exchange and transport phenomena as well.
Professor Dr.-Ing. Ralf Takors
Universität Stuttgart
Institut für Bioverfahrenstechnik (IBVT)
Allmandring 31, 70569 Stuttgart
Dr. Steffen Lindner-Mehlich
Charité Berlin
Professor Dr. Georg Sprenger
Universität Stuttgart
Institut für Mikrobiologie (IMB)
Allmandring 31, 70569 Stuttgart
Sustainable production is becoming increasingly important. Sustainable second-generation biofuel is therefore produced biotechnologically with microorganisms. The project PEBcascade aims to establish a self-sufficient biotechnological reaction cascade for the production of biofuel via ethyl butyrate. In an anaerobic mixed culture, biotechnologically produced Clostridia species are cultivated in combination with yeasts or bacteria. The Clostridia species are responsible for the production of butyrate, and the yeasts or bacteria subsequently produce ethanol in the co-culture cascade. To realize this novel production cascade of a biofuel process developed in the PEBcascade project, expertise in microbiology and molecular biology is combined with biochemical process techniques for process development and modeling and subsequent scale-up to 50 liters. Special enzymes are needed for these next-generation biofuels. Among the most important are lipases. Therefore, different microbial strains and transporter systems will be compared to determine appropriate surface expression of lipase. The performance and kinetics of the autarkic reaction cascade will also be investigated. The data obtained will be used to refine the mathematical model. PEBcascade plans to test bioengineered ethyl butyrate in a single-cylinder gasoline engine at the Institute of Vehicle Propulsion Systems (TUK).
Prof. Dr.-Ing. Roland Ulber, TU Kaiserslautern
Prof. Dr.-Ing. Dirk Holtmann, TH Mittelhessen
Prof. Dr.-Ing Michael Günthner, TU Kaiserslautern

Ralf Takors
Prof. Dr.-Ing.Professor
Martina Rehnert
Dr.Post-Doc, Project Manager DFG SPP 2170 „InterZell“